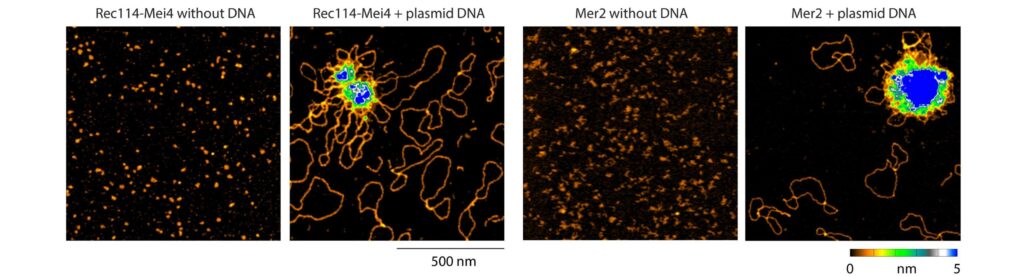
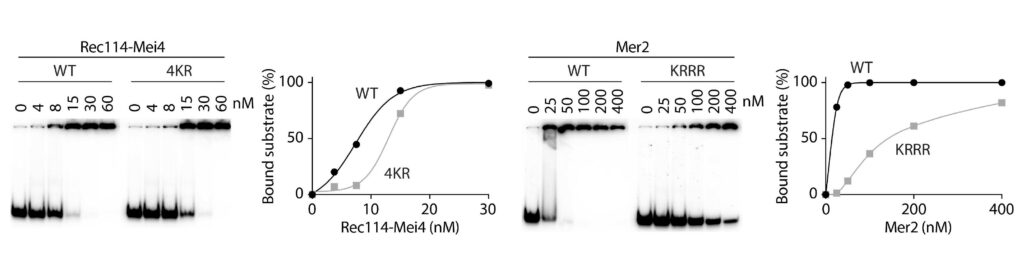
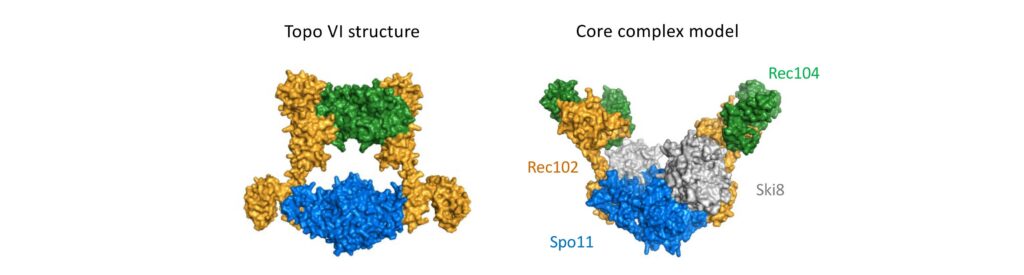
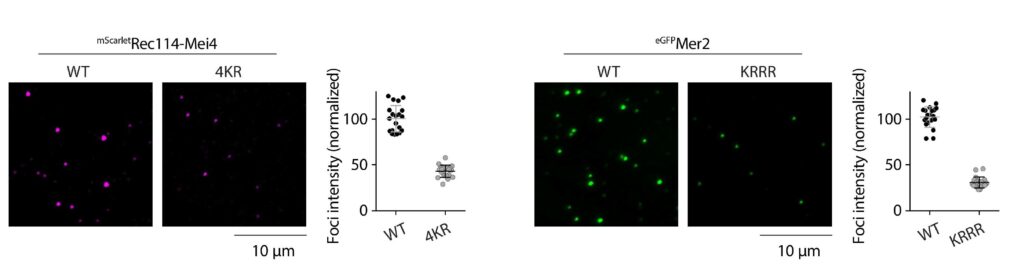
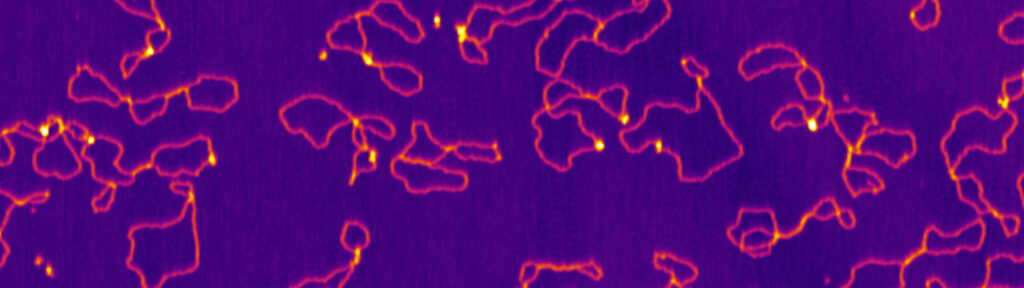
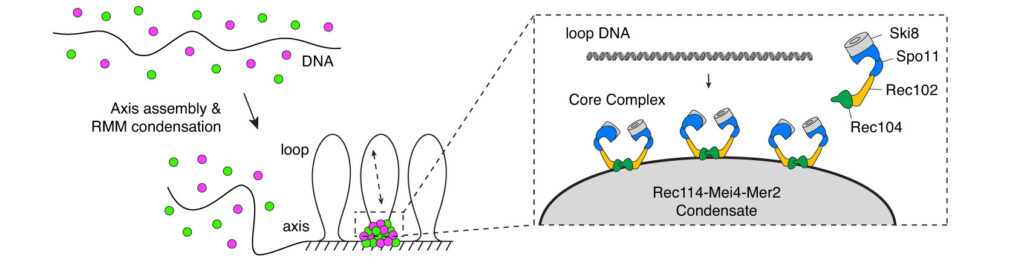
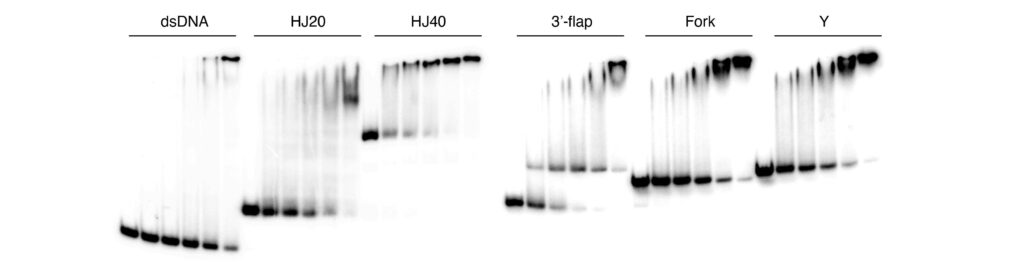
The mechanism of crossover formation
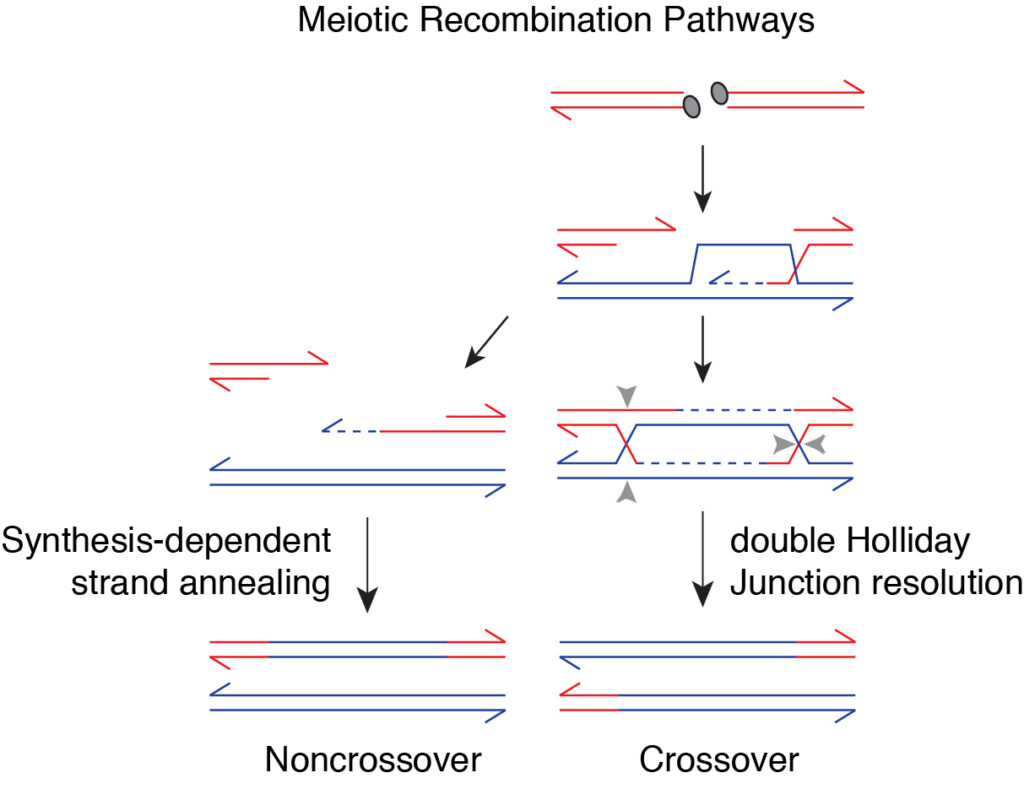
After DSB formation, a broken DNA end invades a homologous sequence which is used as a template to synthesize DNA across the break. A subset of these recombination intermediates mature into double-Holliday junctions (dHJs), which are eventually resolved as crossovers (Figure 6). Other recombination events are repaired through alternative pathways that give primarily non-crossovers.
The specific conversion of dHJs into crossover products implies that the resolution mechanism involves a functional coordination between the two HJ structures. However, the underlying mechanism is mysterious.
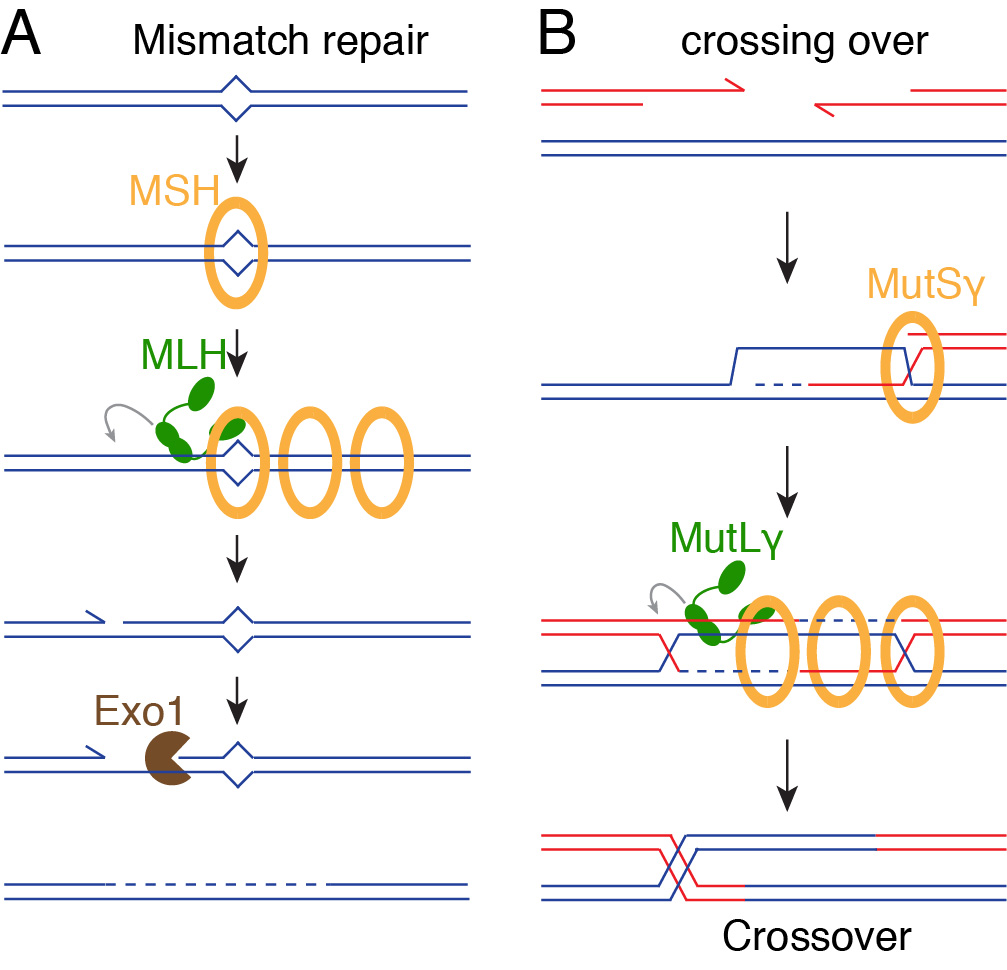
The MutLγ complex, together with its partners MutSγ and Exo1, is thought to resolve dHJs into crossovers. MutLγ and MutSγ are related to mismatch repair (MMR) proteins whose function is to correct errors that arise during DNA replication. During MMR, a MSH heterodimer recognizes a mismatch-containing substrate and recruits a MLH heterodimer, which introduces a single-strand nick (Figure 7A). This provides an entry point for Exo1 that degrades the newly synthesized strand, which allows polymerases to correct the mismatch.
By analogy, it is generally thought that MutSγ recognizes a recombination intermediate and recruits MutLγ, which introduces DNA nicks that lead to the formation of crossovers (Figure 7B). However, this is more of a conceptual mechanism, and the key activities have not been reconstituted. We have purified MutLγ and MutSγ and seek to gain a better understanding of the mechanism of crossover formation.
Recombination and the synaptonemal complex
Crossover formation happens in the context of the higher-order structure of meiotic chromosomes where the two homologs synapse along their entire length through a nucleoprotein structure called the synaptonemal complex (SC) (Figure 8). There is considerable interplay between recombination and the SC, since many proteins that affect crossover formation affect the SC, and vice versa. We also aim to learn more about these structural and functional relationships.

